By remodeling normal H&E slides into digital multiplex protein maps, GigaTIME reveals how immune exercise, tumor invasion, and survival are linked throughout 1000’s of cancers, opening a brand new path for scalable, data-driven oncology analysis.
Research: Multimodal AI generates digital inhabitants for tumor microenvironment modeling
A latest research printed within the journal Cell explored the capabilities of GigaTIME, a multimodal AI framework designed for large-scale modeling of the tumor immune microenvironment (TIME) in most cancers analysis.
TIME Complexity and Profiling Challenges
The TIME is a extremely complicated spatial ecosystem composed of most cancers cells and a wide range of non-malignant cell varieties, reminiscent of cancer-associated fibroblasts (CAFs), immune cells, endothelial cells (ECs), pericytes, and others, all embedded inside a transformed extracellular matrix. It’s profoundly related to most cancers development, shaping tumor progress, invasion, metastasis, and therapeutic outcomes by means of its regulation of immune surveillance and facilitation of immune evasion.
Researchers make use of immunohistochemistry (IHC) to characterize cell states inside the TIME. For instance, PD-L1 IHC staining is used to detect PD-L1 expression, a typical biomarker for predicting response to checkpoint inhibitor therapies.
A significant downside of IHC is that protein activation is assessed individually, requiring a separate tissue pattern for every evaluation. This limitation poses a big problem for modeling the tumor microenvironment, as understanding the intricate interactions between tumor and immune cells requires evaluating a number of proteins concurrently. Multiplex immunofluorescence (mIF) addresses this concern by enabling co-localized, multi-channel protein profiling on the identical tissue part, whereas sustaining spatial group.
Regardless of its promise, mIF is prohibitively costly for large-scale research, requiring expensive reagents, specialised tools, and labor-intensive workflows, thereby limiting dataset availability and medical purposes. In distinction, hematoxylin and eosin (H&E) staining is broadly used and cheap in medical observe to look at tissue and cell morphology. Though H&E pictures don’t straight present cell states, their patterns could trace at them. AI fashions educated on many pathology pictures can detect options linked to the place proteins are energetic in tissue.
AI-Based mostly Digital mIF Technology
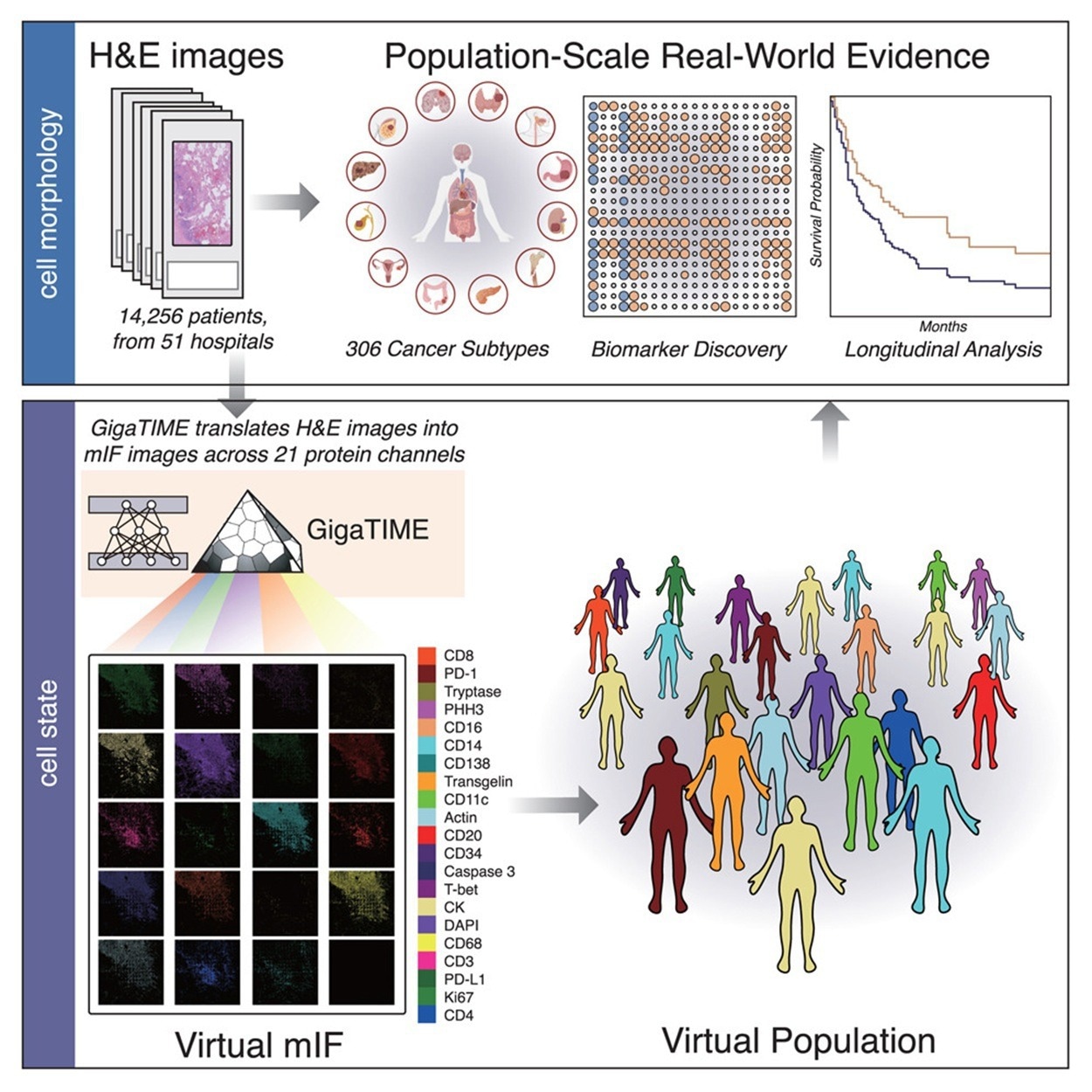
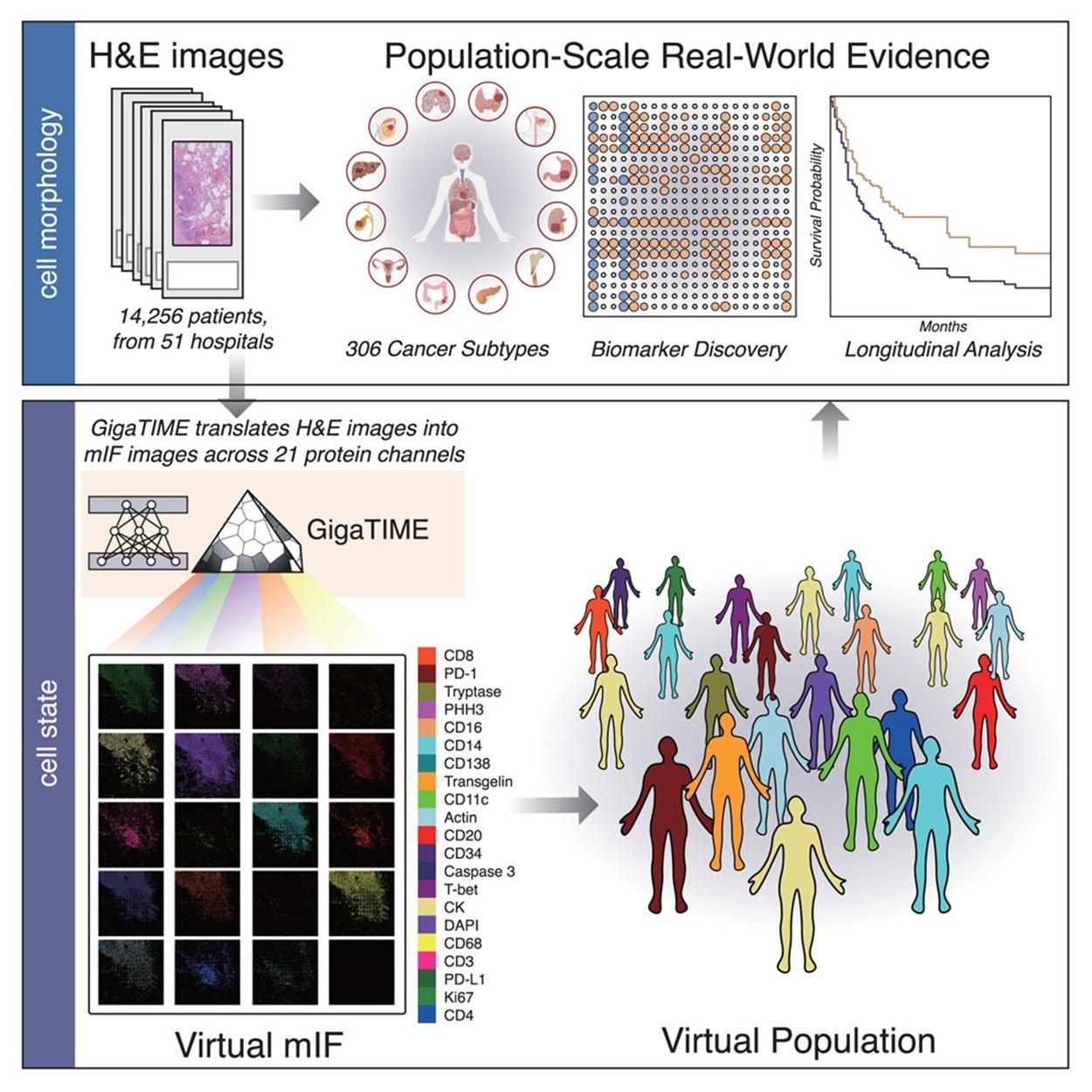
GigaTIME generates various digital mIF populations from large-scale H&E slides. 4 hundred and forty-one mIF pictures had been acquired from 21 H&E-stained slides spanning 21 protein channels to create the coaching dataset. After picture registration and cell segmentation, this yielded a dataset of 40 million matched cells.
GigaTIME was utilized to 14,256 whole-slide H&E pictures from Windfall Well being, which coated 24 most cancers varieties and 306 subtypes throughout 51 hospitals and over 1,000 clinics. The mannequin generated 299,376 digital mIF pictures, leading to a big multimodal dataset with related medical data. The evaluation of the whole-slide H&E pictures produced a complete TIME spectrum and atlas, and revealed greater than 1,200 (particularly, 1,234) vital associations between medical biomarkers and protein channels.
For every digital mIF picture, protein activation density scores had been calculated and, subsequently, aggregated by most cancers subtype to profile mIF-based TIME options. Along with density, the authors quantified spatial metrics reminiscent of entropy, sharpness, and signal-to-noise ratio, which in some circumstances confirmed stronger associations with medical biomarkers than density alone. GigaTIME was additionally utilized to 10,200 TCGA tumors to generate 214,200 digital mIF pictures, thereby demonstrating its robustness. The mannequin’s generalizability and reliability had been confirmed by the robust concordance between datasets in protein activation patterns and biomarker associations.
GigaTIME’s efficiency in translating H&E pictures to mIF pictures was benchmarked in opposition to CycleGAN utilizing three metrics on the pixel, cell, and slide ranges. GigaTIME outperformed CycleGAN on 15 of 21 protein channels, underscoring the worth of paired H&E and mIF information.
Out-of-sample evaluation of GigaTIME’s generalizability was carried out by testing it on breast and mind tumor microarrays that weren’t included within the coaching set. Regardless of various most cancers varieties, phases, and pattern codecs, GigaTIME maintained robust efficiency in Cube scores and correlations, persistently outperforming CycleGAN and baseline strategies throughout most cancers varieties.
Stratified evaluation by subcellular localization confirmed that nuclear proteins had greater translation high quality than floor or cytoplasmic proteins, possible as a result of their compact, well-defined buildings are simpler to foretell. Some cytoplasmic and membrane proteins could also be inherently much less translatable from morphology alone, reflecting elementary limits of H&E-to-protein inference.
Protein Signatures Linked to Tumor Invasion
GigaTIME recognized spatial and combinatorial protein activation patterns and enabled risk-based affected person stratification by stage and survival. Many associations differed by most cancers sort and histological subtype, highlighting the organic heterogeneity of the TIME. Validation utilizing tumors from the Most cancers Genome Atlas (TCGA) supported the generalizability of digital protein activation patterns relatively than straight validating staging predictions.
Research performed on GigaTIME digital populations documented that, on the pan-cancer stage, tumor invasion stage was related to elevated digital PD-L1 activation and complicated patterns of protein activation. This mirrored a coordinated immune response. In superior illness, the information instructed that various immune evasion mechanisms turned more and more influential over PD-L1-mediated pathways. The doable evasion of immune-induced apoptosis was indicated by lowered predicted cleaved caspase-3 expression.
Protein activations for a number of immune cell markers confirmed robust cross-correlation, supporting the necessity for immunotherapies concentrating on numerous cell varieties. The GigaTIME signature, combining all protein channels, outperformed single-channel fashions in predicting survival. Associations had been discovered between GigaTIME digital protein activations and each identified and fewer well-described genomic alterations, in addition to decreased immunogenicity linked to oncogene mutations reminiscent of KRAS.
For instance, the authors spotlight combinatorial relationships reminiscent of CD138 with CD68, and PD-L1 with cleaved caspase 3, illustrating how spatial and multiprotein signatures can reveal immune-tumor interactions not evident from single markers alone.
Increasing Entry to Spatial Proteomics
Preliminary outcomes demonstrated GigaTIME’s promise, with the most important digital mIF affiliation research to this point. By enabling population-scale, spatially resolved proteomic inference from routine H&E slides, the method has the potential to broaden entry to detailed tumor immune profiling in each analysis and medical settings. Nonetheless, there’s a want for extra geographic and ethnic variety, as most sufferers had been from the western United States.
The findings confirmed that H&E slides seize vital spatial proteomic alerts, however translation high quality assorted throughout protein channels. This variability may stem from numerous components, together with assorted tissue structure, variations in underlying coaching information, organic heterogeneity, and marker-specific technical challenges. The authors word that sure proteins could also be inconceivable to precisely infer from H&E morphology, underscoring the sensible limits of digital mIF.
Ongoing work goals to evaluate extra protein channels, assemble a complete digital mIF atlas, and incorporate cell segmentation fashions to shed extra mild on cell-to-cell interactions within the tumor microenvironment.
Leave a Reply